How do I import my data?
Table of contents
All data, with the exception of genotypic data, can be imported into your site using friendly web-based forms!
Web-based Data Import Forms
- To import data, go to “Tripal” on the top administration toolbar. The click on “Data Loaders”.
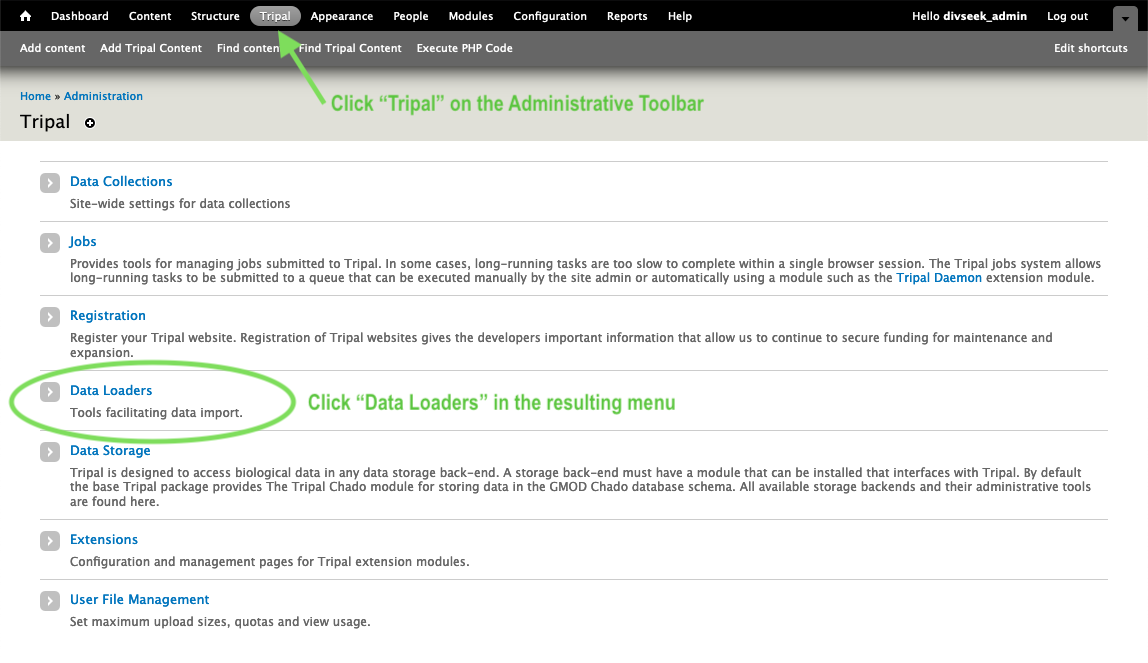
- Then choose your data type from the list of Data Loaders. For example, for germplasm accession passport data you would choose “Germplasm Accession Importer”.
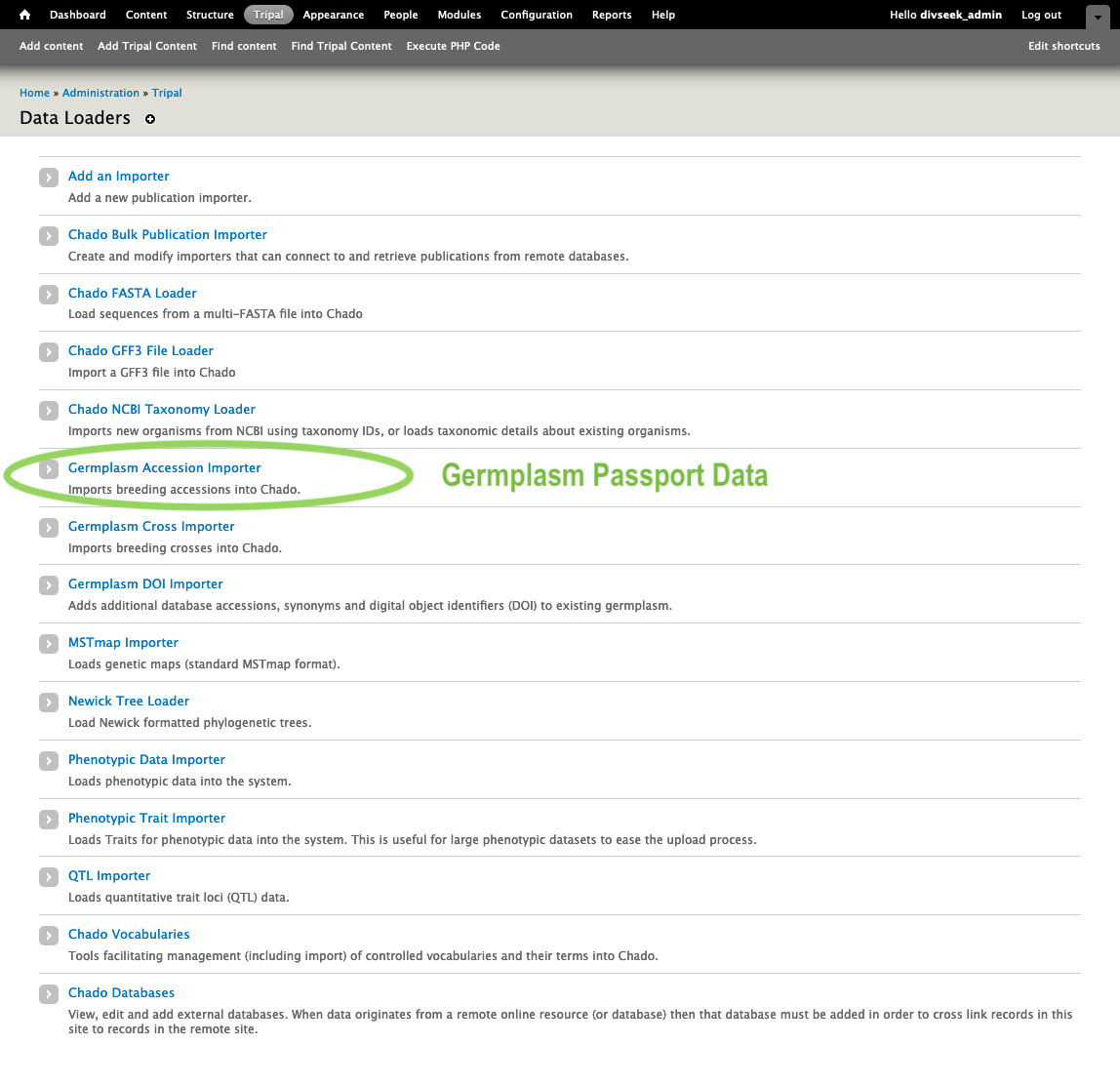
- Each data importer provides detailed instructions on the format of file needed to import that data type. Where there are clear standards (e.g VCF or MSTmap) they are used but in many cases there are not yet good standards (e.g. quantitative trait loci). Here is the germplasm importer instructions.
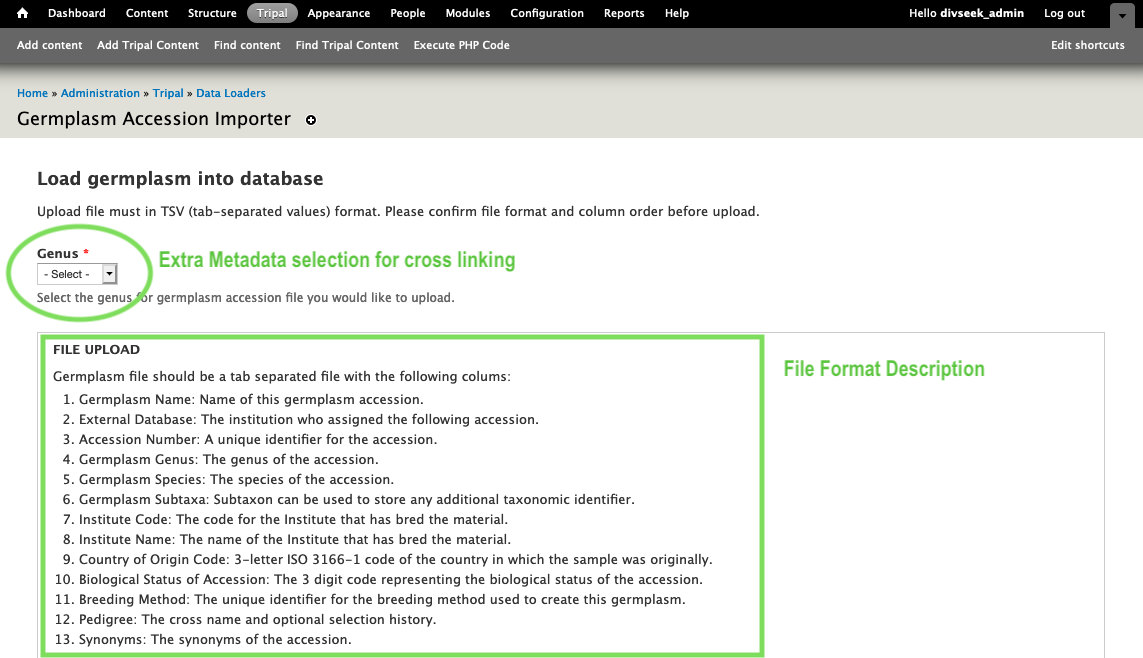
- Once you have your data prepared in the described format, you can use the Browse button to select your file and then the upload button to upload it to your site. Then you just fill out any additional metadata and click “Import file” to submit the job!
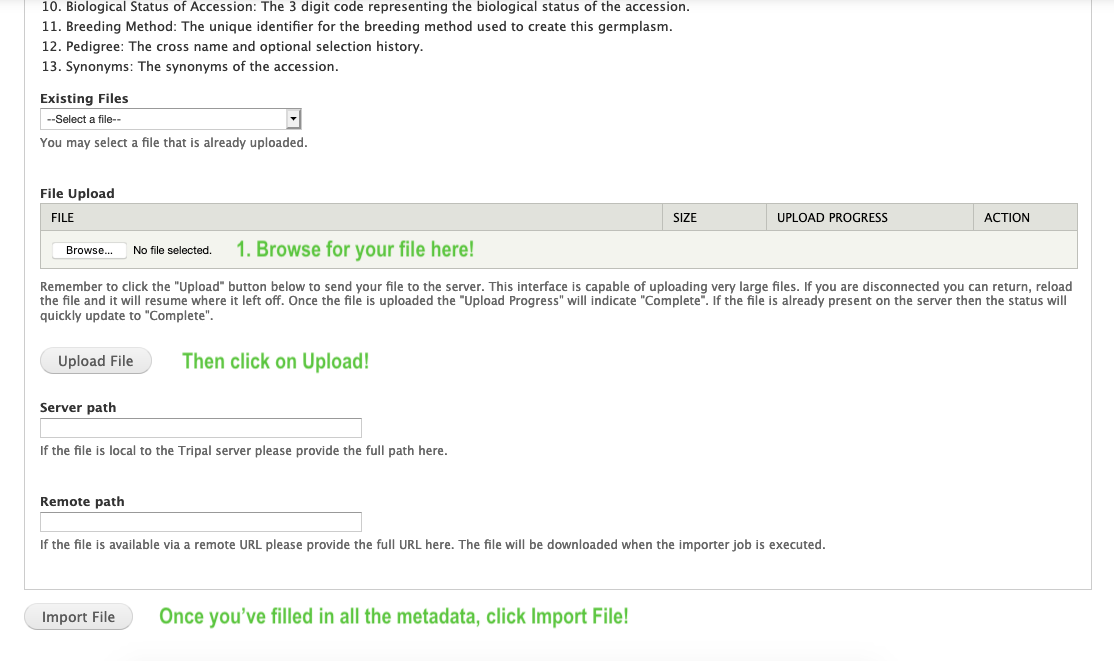
- Since biological data files can be large, all data is imported from the command line. When you submitted the job above, you should have seen a green message giving you the command to run on the command-line!
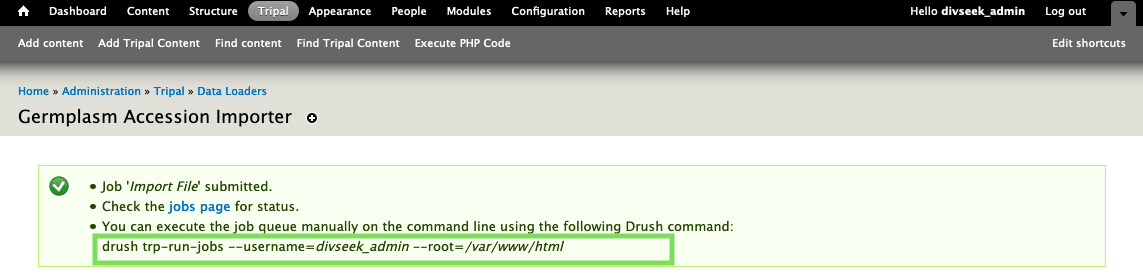
To do this, you first need to SSH into your server. For the ComputeCanada OpenStack cloud, you can find instructions on step 10. Then execute the command inside your site docker container as follows:
sudo docker exec -it --workdir=/var/www/html tcrop vendor/bin/drush trp-run-jobs --username=divseek_admin --root=/var/www/html
Make sure to change the username to match your administrative user as shown in the screenshot for this step!
Genotypic Data
Genotypic Data is fully imported on the command-line. There are detailed instructions on the Tripal Genotypes Loader extension module documentation.
Phenotypic Data
The web-based for for phenotypic data has multiple steps as it validates data before importing it. You can find detailed instructions on the Tripal Analyzed Phenotypes extension module documentation.